| Revision as of 09:26, 11 March 2013 edit211.154.201.34 (talk) deleted the content of purely advertisements which were obviously from the authors, and only keep those truly well-known methods in the field.← Previous edit | Revision as of 05:56, 14 March 2013 edit undo72.227.168.130 (talk) Undid revision 543381008 by 211.154.201.34 (talk)Next edit → | ||
| Line 57: | Line 57: | ||
| The ] has recently released an updated version of CE and FATCAT as part of the . It provides a new variation of CE that can detect ] in protein structures.<ref name="prlic"/> | The ] has recently released an updated version of CE and FATCAT as part of the . It provides a new variation of CE that can detect ] in protein structures.<ref name="prlic"/> | ||
| ===GANGSTA+=== | |||
| GANGSTA+ is a combinatorial algorithm for non-sequential structural alignment of proteins and similarity search in databases.<ref name="GuerlerKnapp"/> | |||
| It uses a combinatorial approach on the secondary structure level to evaluate similarities between two protein structures based on contact maps. Different SSE assignment modes can be used. The assignment of SSEs can be performed respecting the sequential order of the SSEs in the polypeptide chains of the considered protein pair (sequential alignment) or by ignoring this order (non-sequential alignment). Furthermore, SSE pairs can optionally be aligned in reverse orientation. | |||
| The highest ranking SSE assignments are transferred to the residue level by a pointmatching approach.<ref name="Guerler"/> To obtain an initial common set of atomic coordinates for both proteins, pairwise attractive interactions of the C-alpha atom pairs are defined by inverse Lorentzians and energy minimized. | |||
| ===MAMMOTH=== | |||
| '''MA'''tching '''M'''olecular '''M'''odels '''O'''btained from '''TH'''eory. As its name suggests, MAMMOTH was originally developed for comparing models coming from structure prediction ('''TH'''eory) since it is tolerant of large unalignable regions, but it has proven to work well with experimental models, especially when looking for remote homology. Benchmarks on targets of blind structure prediction (the ] experiment) and automated GO annotation have shown it is tightly rank correlated with human curated annotation.<ref name="ortiz" /><ref name="svm_go_annotation"/> A highly complete database of mammoth-based structure annotations for the predicted structures of unknown proteins covering 150 genomes facilitates genomic scale normalization.<ref name='wcg_hpf1'>. Predictions of structures of all unknown small domains in 150 genones. nyu.edu</ref> | |||
| '''MAMMOTH'''-based structure alignment methods decompose the protein structure into short, seven-residue, peptides (heptapeptides) which are compared with the heptapeptides of another protein. The similarity score between two heptapeptides is calculated using a unit-vector RMS (URMS) method.<ref name=kedem/> These scores are stored in a similarity matrix, and with a hybrid (local-global) ] the optimal residue alignment is calculated. Protein similarity scores calculated with MAMMOTH is derived from the likelihood of obtaining a given structural alignment by chance.<ref name="ortiz"/> | |||
| '''MAMMOTH-mult''' is an extension of the MAMMOTH algorithm to be used to align related families of protein structures. This algorithm is very fast and produces consistent and high quality structural alignments.<ref name=lupyan/> Multiple structural alignments calculated with MAMMOTH-mult produces structurally implied sequence alignments that can be further used for multiple-template homology modeling, ]-based ], and profile-type PSI-BLAST searches. | |||
| ===ProBiS=== | |||
| '''Pro'''tein '''Bi'''nding '''S'''ites. '''ProBiS''' detects structurally similar sites on protein surfaces by local surface structure alignment. It compares the query protein to members of a database of protein 3D structures and detects with sub-residue precision, structurally similar sites as patterns of physicochemical properties on the protein surface. Using an efficient maximum clique algorithm, the program identifies proteins that share local structural similarities with the query protein and generates structure-based alignments of these proteins with the query. Structural similarity scores are calculated for the query protein’s surface residues, and are expressed as different colors on the query protein surface. The algorithm has been used successfully for the detection of protein–protein, protein–small ligand and protein–DNA binding sites.<ref name="Konc"/> | |||
| ===RAPIDO=== | |||
| '''R'''apid '''A'''lignment of '''P'''roteins '''I'''n terms of '''DO'''mains. '''RAPIDO''' is a web server for the 3D alignment of crystal structures of different protein molecules, in the presence of conformational changes.<ref name="mosca"/> Similar to what CE does as a first step, RAPIDO identifies fragments that are structurally similar in the two proteins using an approach based on difference distance matrices. The Matching Fragment Pairs (MFPs) are then represented as nodes in a graph which are chained together to form an alignment by means of an algorithm for the identification of the ] on a ]. The final step of refinement is performed to improve the quality of the alignment. After aligning the two structures the server applies a genetic algorithm for the identification of conformationally invariant regions.<ref name="schneider"/> These regions correspond to groups of atoms whose interatomic distances are constant (within a defined tolerance). In doing so RAPIDO takes into account the variation in the reliability of atomic coordinates by employing weighting-functions based on the refined B-values. The regions identified as conformationally invariant by RAPIDO represent reliable sets of atoms for the superposition of the two structures that can be used for a detailed analysis of changes in the conformation. In addition to the functionalities provided by existing tools, RAPIDO can identify structurally equivalent regions even when these consist of fragments that are distant in terms of sequence and separated by other movable domains. | |||
| ===SABERTOOTH=== | |||
| SABERTOOTH uses structural profiles to perform structural alignments. The underlying structural profiles expresses the global connectivity of each residue. Despite the very condensed vectorial representation, the tool recognizes structural similarities with accuracy comparable to established alignment tools based on coordinates and performs comparably in quality.<ref name="SABERTOOTH"/> Furthermore, the algorithm has favourable scaling of computation time with chain length. Since the algorithm is independent of the details of the structural representation, the framework can be generalized to sequence-to-sequence and sequence-to-structure comparison within the same setup, and it is therefore more general than other tools. | |||
| SABERTOOTH can be used online at | |||
| ===SSAP=== | ===SSAP=== | ||
| Line 62: | Line 84: | ||
| SSAP originally produced only pairwise alignments but has since been extended to multiple alignments as well.<ref name="taylor"/> It has been applied in an all-to-all fashion to produce a hierarchical fold classification scheme known as ] (Class, Architecture, Topology, Homology),<ref name="orengo"/> which has been used to construct the database. | SSAP originally produced only pairwise alignments but has since been extended to multiple alignments as well.<ref name="taylor"/> It has been applied in an all-to-all fashion to produce a hierarchical fold classification scheme known as ] (Class, Architecture, Topology, Homology),<ref name="orengo"/> which has been used to construct the database. | ||
| ===SPalign=== | |||
| SPalign is based on a new size-independent score called SPscore for pairwise protein structure alignment. SP-score fixes the cutoff distance at 4 Å and removed the size dependence using a normalization pre-factor. The method is compared to DALI, CE, TMalign and others in fold recognition and function prediction.<ref name="yang"/> The source code for SPalign and the server are available at http://sparks.informatics.iupui.edu/yueyang/server/SPalign/. | |||
| ===TOPOFIT=== | |||
| In the TOPOFIT method,<ref>{{cite web | title=Protein structure alignment by TOPOFIT | author=Ilyin lab | url=http://topofit.ilyinlab.org/topofit/index.php similarity}}</ref> of protein structures is analyzed using three-dimensional Delaunay triangulation patterns derived from backbone representation. It has been found that structurally related proteins have a common spatial invariant part, a set of tetrahedrons, mathematically described as a common spatial sub-graph volume of the three-dimensional contact graph derived from Delaunay tessellation (DT). Based on this property of protein structures we present a novel common volume superimposition (TOPOFIT) method to produce structural alignments of proteins. The superimposition of the DT patterns allows one to objectively identify a common number of equivalent residues in the structural alignment, in other words, TOPOFIT identifies a feature point on the RMSD/Ne curve, a topomax point, until which two structures correspond to each other including backbone and inter-residue contacts, while the growing number of mismatches between the DT patterns occurs at larger RMSD (Ne) after topomax point. The topomax point is present in all alignments from different protein structural classes; therefore, the TOPOFIT method identifies common, invariant structural parts between proteins. The TOPOFIT method adds new opportunities for the comparative analysis of protein structures and for more detailed studies on understanding the molecular principles of tertiary structure organization and functionality. It helps to detect conformational changes, topological differences in variable parts, which are particularly important for studies of variations in active/binding sites and protein classification.<ref name="Valentin"/> | |||
| ===SSM=== | |||
| Secondary Structure Matching (SSM),<ref name="SSM"/> or at the Protein Data Bank in Europe uses graph matching followed by c-alpha alignment to compute alignments. | |||
| ==Recent developments== | ==Recent developments== | ||
Revision as of 05:56, 14 March 2013
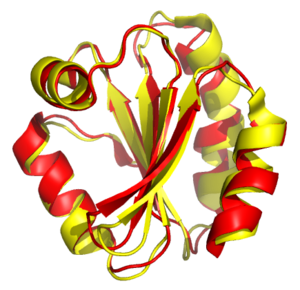
Structural alignment attempts to establish homology between two or more polymer structures based on their shape and three-dimensional conformation. This process is usually applied to protein tertiary structures but can also be used for large RNA molecules. In contrast to simple structural superposition, where at least some equivalent residues of the two structures are known, structural alignment requires no a priori knowledge of equivalent positions. Structural alignment is a valuable tool for the comparison of proteins with low sequence similarity, where evolutionary relationships between proteins cannot be easily detected by standard sequence alignment techniques. Structural alignment can therefore be used to imply evolutionary relationships between proteins that share very little common sequence. However, caution should be used in using the results as evidence for shared evolutionary ancestry because of the possible confounding effects of convergent evolution by which multiple unrelated amino acid sequences converge on a common tertiary structure.
Structural alignments can compare two sequences or multiple sequences. Because these alignments rely on information about all the query sequences' three-dimensional conformations, the method can only be used on sequences where these structures are known. These are usually found by X-ray crystallography or NMR spectroscopy. It is possible to perform a structural alignment on structures produced by structure prediction methods. Indeed, evaluating such predictions often requires a structural alignment between the model and the true known structure to assess the model's quality. Structural alignments are especially useful in analyzing data from structural genomics and proteomics efforts, and they can be used as comparison points to evaluate alignments produced by purely sequence-based bioinformatics methods.
The outputs of a structural alignment are a superposition of the atomic coordinate sets and a minimal root mean square deviation (RMSD) between the structures. The RMSD of two aligned structures indicates their divergence from one another. Structural alignment can be complicated by the existence of multiple protein domains within one or more of the input structures, because changes in relative orientation of the domains between two structures to be aligned can artificially inflate the RMSD.
Data produced by structural alignment
The minimum information produced from a successful structural alignment is a set of superposed three-dimensional coordinates for each input structure. (Note that one input element may be fixed as a reference and therefore its superposed coordinates do not change.) The fitted structures can be used to calculate mutual RMSD values, as well as other more sophisticated measures of structural similarity such as the global distance test (GDT, the metric used in CASP). The structural alignment also implies a corresponding one-dimensional sequence alignment from which a sequence identity, or the percentage of residues that are identical between the input structures, can be calculated as a measure of how closely the two sequences are related.
Types of comparisons
Because protein structures are composed of amino acids whose side chains are linked by a common protein backbone, a number of different possible subsets of the atoms that make up a protein macromolecule can be used in producing a structural alignment and calculating the corresponding RMSD values. When aligning structures with very different sequences, the side chain atoms generally are not taken into account because their identities differ between many aligned residues. For this reason it is common for structural alignment methods to use by default only the backbone atoms included in the peptide bond. For simplicity and efficiency, often only the alpha carbon positions are considered, since the peptide bond has a minimally variant planar conformation. Only when the structures to be aligned are highly similar or even identical is it meaningful to align side-chain atom positions, in which case the RMSD reflects not only the conformation of the protein backbone but also the rotameric states of the side chains. Other comparison criteria that reduce noise and bolster positive matches include secondary structure assignment, native contact maps or residue interaction patterns, measures of side chain packing, and measures of hydrogen bond retention.
Structural superposition
The most basic possible comparison between protein structures makes no attempt to align the input structures and requires a precalculated alignment as input to determine which of the residues in the sequence are intended to be considered in the RMSD calculation. Structural superposition is commonly used to compare multiple conformations of the same protein (in which case no alignment is necessary, since the sequences are the same) and to evaluate the quality of alignments produced using only sequence information between two or more sequences whose structures are known. This method traditionally uses a simple least-squares fitting algorithm, in which the optimal rotations and translations are found by minimizing the sum of the squared distances among all structures in the superposition. More recently, maximum likelihood and Bayesian methods have greatly increased the accuracy of the estimated rotations, translations, and covariance matrices for the superposition.
Algorithms based on multidimensional rotations and modified quaternions have been developed to identify topological relationships between protein structures without the need for a predetermined alignment. Such algorithms have successfully identified canonical folds such as the four-helix bundle. The SuperPose method is sufficiently extensible to correct for relative domain rotations and other structural pitfalls.
Algorithmic complexity
Optimal solution
The optimal "threading" of a protein sequence onto a known structure and the production of an optimal multiple sequence alignment have been shown to be NP-complete. However, this does not imply that the structural alignment problem is NP-complete. Strictly speaking, an optimal solution to the protein structure alignment problem is only known for certain protein structure similarity measures, such as the measures used in protein structure prediction experiments, GDT_TS and MaxSub. These measures can be rigorously optimized using an algorithm capable of maximizing the number of atoms in two proteins that can be superimposed under a predefined distance cutoff. Unfortunately, the algorithm for optimal solution is not practical, since its running time depends not only on the lengths but also on the intrinsic geometry of input proteins.
Approximate solution
Approximate polynomial-time algorithms for structural alignment that produce a family of "optimal" solutions within an approximation parameter for a given scoring function have been developed. Although these algorithms theoretically classify the approximate protein structure alignment problem as "tractable", they are still computationally too expensive for large-scale protein structure analysis. As a consequence, practical algorithms that converge to the global solutions of the alignment, given a scoring function, do not exist. Most algorithms are, therefore, heuristic, but algorithms that guarantee the convergence to at least local maximizers of the scoring functions, and are practical, have been developed.
Representation of structures
Protein structures have to be represented in some coordinate-independent space to make them comparable. This is typically achieved by constructing a sequence-to-sequence matrix or series of matrices that encompass comparative metrics: rather than absolute distances relative to a fixed coordinate space. An intuitive representation is the distance matrix, which is a two-dimensional matrix containing all pairwise distances between some subset of the atoms in each structure (such as the alpha carbons). The matrix increases in dimensionality as the number of structures to be simultaneously aligned increases. Reducing the protein to a coarse metric such as secondary structure elements (SSEs) or structural fragments can also produce sensible alignments, despite the loss of information from discarding distances, as noise is also discarded. Choosing a representation to facilitate computation is critical to developing an efficient alignment mechanism.
Methods
Structural alignment techniques have been used in comparing individual structures or sets of structures as well as in the production of "all-to-all" comparison databases that measure the divergence between every pair of structures present in the Protein Data Bank (PDB). Such databases are used to classify proteins by their fold.
DALI

A common and popular structural alignment method is the DALI, or distance alignment matrix method, which breaks the input structures into hexapeptide fragments and calculates a distance matrix by evaluating the contact patterns between successive fragments. Secondary structure features that involve residues that are contiguous in sequence appear on the matrix's main diagonal; other diagonals in the matrix reflect spatial contacts between residues that are not near each other in the sequence. When these diagonals are parallel to the main diagonal, the features they represent are parallel; when they are perpendicular, their features are antiparallel. This representation is memory-intensive because the features in the square matrix are symmetrical (and thus redundant) about the main diagonal.
When two proteins' distance matrices share the same or similar features in approximately the same positions, they can be said to have similar folds with similar-length loops connecting their secondary structure elements. DALI's actual alignment process requires a similarity search after the two proteins' distance matrices are built; this is normally conducted via a series of overlapping submatrices of size 6x6. Submatrix matches are then reassembled into a final alignment via a standard score-maximization algorithm — the original version of DALI used a Monte Carlo simulation to maximize a structural similarity score that is a function of the distances between putative corresponding atoms. In particular, more distant atoms within corresponding features are exponentially downweighted to reduce the effects of noise introduced by loop mobility, helix torsions, and other minor structural variations. Because DALI relies on an all-to-all distance matrix, it can account for the possibility that structurally aligned features might appear in different orders within the two sequences being compared.
The DALI method has also been used to construct a database known as FSSP (Fold classification based on Structure-Structure alignment of Proteins, or Families of Structurally Similar Proteins) in which all known protein structures are aligned with each other to determine their structural neighbors and fold classification. There is an searchable database based on DALI as well as a downloadable program and web search based on a standalone version known as DaliLite.
Combinatorial extension
The combinatorial extension (CE) method is similar to DALI in that it too breaks each structure in the query set into a series of fragments that it then attempts to reassemble into a complete alignment. A series of pairwise combinations of fragments called aligned fragment pairs, or AFPs, are used to define a similarity matrix through which an optimal path is generated to identify the final alignment. Only AFPs that meet given criteria for local similarity are included in the matrix as a means of reducing the necessary search space and thereby increasing efficiency. A number of similarity metrics are possible; the original definition of the CE method included only structural superpositions and inter-residue distances but has since been expanded to include local environmental properties such as secondary structure, solvent exposure, hydrogen-bonding patterns, and dihedral angles.
An alignment path is calculated as the optimal path through the similarity matrix by linearly progressing through the sequences and extending the alignment with the next possible high-scoring AFP pair. The initial AFP pair that nucleates the alignment can occur at any point in the sequence matrix. Extensions then proceed with the next AFP that meets given distance criteria restricting the alignment to low gap sizes. The size of each AFP and the maximum gap size are required input parameters but are usually set to empirically determined values of 8 and 30 respectively. Like DALI and SSAP, CE has been used to construct an all-to-all fold classification database from the known protein structures in the PDB.
The RCSB PDB has recently released an updated version of CE and FATCAT as part of the RCSB PDB Protein Comparison Tool. It provides a new variation of CE that can detect circular permutations in protein structures.
GANGSTA+
GANGSTA+ is a combinatorial algorithm for non-sequential structural alignment of proteins and similarity search in databases. It uses a combinatorial approach on the secondary structure level to evaluate similarities between two protein structures based on contact maps. Different SSE assignment modes can be used. The assignment of SSEs can be performed respecting the sequential order of the SSEs in the polypeptide chains of the considered protein pair (sequential alignment) or by ignoring this order (non-sequential alignment). Furthermore, SSE pairs can optionally be aligned in reverse orientation. The highest ranking SSE assignments are transferred to the residue level by a pointmatching approach. To obtain an initial common set of atomic coordinates for both proteins, pairwise attractive interactions of the C-alpha atom pairs are defined by inverse Lorentzians and energy minimized.
MAMMOTH
MAtching Molecular Models Obtained from THeory. As its name suggests, MAMMOTH was originally developed for comparing models coming from structure prediction (THeory) since it is tolerant of large unalignable regions, but it has proven to work well with experimental models, especially when looking for remote homology. Benchmarks on targets of blind structure prediction (the CASP experiment) and automated GO annotation have shown it is tightly rank correlated with human curated annotation. A highly complete database of mammoth-based structure annotations for the predicted structures of unknown proteins covering 150 genomes facilitates genomic scale normalization.
MAMMOTH-based structure alignment methods decompose the protein structure into short, seven-residue, peptides (heptapeptides) which are compared with the heptapeptides of another protein. The similarity score between two heptapeptides is calculated using a unit-vector RMS (URMS) method. These scores are stored in a similarity matrix, and with a hybrid (local-global) dynamic programming the optimal residue alignment is calculated. Protein similarity scores calculated with MAMMOTH is derived from the likelihood of obtaining a given structural alignment by chance.
MAMMOTH-mult is an extension of the MAMMOTH algorithm to be used to align related families of protein structures. This algorithm is very fast and produces consistent and high quality structural alignments. Multiple structural alignments calculated with MAMMOTH-mult produces structurally implied sequence alignments that can be further used for multiple-template homology modeling, HMM-based protein structure prediction, and profile-type PSI-BLAST searches.
ProBiS
Protein Binding Sites. ProBiS detects structurally similar sites on protein surfaces by local surface structure alignment. It compares the query protein to members of a database of protein 3D structures and detects with sub-residue precision, structurally similar sites as patterns of physicochemical properties on the protein surface. Using an efficient maximum clique algorithm, the program identifies proteins that share local structural similarities with the query protein and generates structure-based alignments of these proteins with the query. Structural similarity scores are calculated for the query protein’s surface residues, and are expressed as different colors on the query protein surface. The algorithm has been used successfully for the detection of protein–protein, protein–small ligand and protein–DNA binding sites.
RAPIDO
Rapid Alignment of Proteins In terms of DOmains. RAPIDO is a web server for the 3D alignment of crystal structures of different protein molecules, in the presence of conformational changes. Similar to what CE does as a first step, RAPIDO identifies fragments that are structurally similar in the two proteins using an approach based on difference distance matrices. The Matching Fragment Pairs (MFPs) are then represented as nodes in a graph which are chained together to form an alignment by means of an algorithm for the identification of the longest path on a directed acyclic graph. The final step of refinement is performed to improve the quality of the alignment. After aligning the two structures the server applies a genetic algorithm for the identification of conformationally invariant regions. These regions correspond to groups of atoms whose interatomic distances are constant (within a defined tolerance). In doing so RAPIDO takes into account the variation in the reliability of atomic coordinates by employing weighting-functions based on the refined B-values. The regions identified as conformationally invariant by RAPIDO represent reliable sets of atoms for the superposition of the two structures that can be used for a detailed analysis of changes in the conformation. In addition to the functionalities provided by existing tools, RAPIDO can identify structurally equivalent regions even when these consist of fragments that are distant in terms of sequence and separated by other movable domains.
SABERTOOTH
SABERTOOTH uses structural profiles to perform structural alignments. The underlying structural profiles expresses the global connectivity of each residue. Despite the very condensed vectorial representation, the tool recognizes structural similarities with accuracy comparable to established alignment tools based on coordinates and performs comparably in quality. Furthermore, the algorithm has favourable scaling of computation time with chain length. Since the algorithm is independent of the details of the structural representation, the framework can be generalized to sequence-to-sequence and sequence-to-structure comparison within the same setup, and it is therefore more general than other tools. SABERTOOTH can be used online at http://www.fkp.tu-darmstadt.de/sabertooth/
SSAP
The SSAP (Sequential Structure Alignment Program) method uses double dynamic programming to produce a structural alignment based on atom-to-atom vectors in structure space. Instead of the alpha carbons typically used in structural alignment, SSAP constructs its vectors from the beta carbons for all residues except glycine, a method which thus takes into account the rotameric state of each residue as well as its location along the backbone. SSAP works by first constructing a series of inter-residue distance vectors between each residue and its nearest non-contiguous neighbors on each protein. A series of matrices are then constructed containing the vector differences between neighbors for each pair of residues for which vectors were constructed. Dynamic programming applied to each resulting matrix determines a series of optimal local alignments which are then summed into a "summary" matrix to which dynamic programming is applied again to determine the overall structural alignment.
SSAP originally produced only pairwise alignments but has since been extended to multiple alignments as well. It has been applied in an all-to-all fashion to produce a hierarchical fold classification scheme known as CATH (Class, Architecture, Topology, Homology), which has been used to construct the CATH Protein Structure Classification database.
SPalign
SPalign is based on a new size-independent score called SPscore for pairwise protein structure alignment. SP-score fixes the cutoff distance at 4 Å and removed the size dependence using a normalization pre-factor. The method is compared to DALI, CE, TMalign and others in fold recognition and function prediction. The source code for SPalign and the server are available at http://sparks.informatics.iupui.edu/yueyang/server/SPalign/.
TOPOFIT
In the TOPOFIT method, of protein structures is analyzed using three-dimensional Delaunay triangulation patterns derived from backbone representation. It has been found that structurally related proteins have a common spatial invariant part, a set of tetrahedrons, mathematically described as a common spatial sub-graph volume of the three-dimensional contact graph derived from Delaunay tessellation (DT). Based on this property of protein structures we present a novel common volume superimposition (TOPOFIT) method to produce structural alignments of proteins. The superimposition of the DT patterns allows one to objectively identify a common number of equivalent residues in the structural alignment, in other words, TOPOFIT identifies a feature point on the RMSD/Ne curve, a topomax point, until which two structures correspond to each other including backbone and inter-residue contacts, while the growing number of mismatches between the DT patterns occurs at larger RMSD (Ne) after topomax point. The topomax point is present in all alignments from different protein structural classes; therefore, the TOPOFIT method identifies common, invariant structural parts between proteins. The TOPOFIT method adds new opportunities for the comparative analysis of protein structures and for more detailed studies on understanding the molecular principles of tertiary structure organization and functionality. It helps to detect conformational changes, topological differences in variable parts, which are particularly important for studies of variations in active/binding sites and protein classification.
SSM
Secondary Structure Matching (SSM), or PDBeFold at the Protein Data Bank in Europe uses graph matching followed by c-alpha alignment to compute alignments.
Recent developments
Improvements in structural alignment methods constitute an active area of research, and new or modified methods are often proposed that are claimed to offer advantages over the older and more widely distributed techniques. A recent example, TM-align, uses a novel method for weighting its distance matrix, to which standard dynamic programming is then applied. The weighting is proposed to accelerate the convergence of dynamic programming and correct for effects arising from alignment lengths. In a benchmarking study, TM-align has been reported to improve in both speed and accuracy over DALI and CE.
However, as algorithmic improvements and computer performance have erased purely technical deficiencies in older approaches, it has become clear that there is no one universal criterion for the 'optimal' structural alignment. TM-align, for instance, is particularly robust in quantifying comparisons between sets of proteins with great disparities in sequence lengths, but it only indirectly captures hydrogen bonding or secondary structure order conservation which might be better metrics for alignment of evolutionarily related proteins. Thus recent developments have focused on optimizing particular attributes such as speed, quantification of scores, correlation to alternative gold standards, or tolerance of imperfection in structural data or ab initio structural models. An alternative methodology that is gaining popularity is to use the consensus of various methods to ascertain proteins structural similarities.
| This section needs expansion with: add discussion of the following topics: A) flexible alignment vs. rigid body B) sequence order dependent vs. independent C) alignment of biological assemblies. You can help by adding to it. (July 2012) |
RNA structural alignment
Structural alignment techniques have traditionally been applied exclusively to proteins, as the primary biological macromolecules that assume characteristic three-dimensional structures. However, large RNA molecules also form characteristic tertiary structures, which are mediated primarily by hydrogen bonds formed between base pairs as well as base stacking. Functionally similar noncoding RNA molecules can be especially difficult to extract from genomics data because structure is more strongly conserved than sequence in RNA as well as in proteins, and the more limited alphabet of RNA decreases the information content of any given nucleotide at any given position.
However, because of the increasing interest in RNA structures and because of the growth of the number of experimentally determined 3D RNA structures, few RNA structure similarity methods have been developed recently. One of those methods is, e.g., SETTER which decomposes each RNA structure into smaller parts called general secondary structure units (GSSUs). GSSUs are subsequently aligned and these partial alignments are merged into the final RNA structure alignment and scored. The method has been implemented into the SETTER webserver.
A recent method for pairwise structural alignment of RNA sequences with low sequence identity has been published and implemented in the program FOLDALIGN. However, this method is not truly analogous to protein structural alignment techniques because it computationally predicts the structures of the RNA input sequences rather than requiring experimentally determined structures as input. Although computational prediction of the protein folding process has not been particularly successful to date, RNA structures without pseudoknots can often be sensibly predicted using free energy-based scoring methods that account for base pairing and stacking.
Software
Main article: Structural alignment softwareChoosing a software tool for structural alignment can be a challenge due to the large variety of available packages that differ significantly in methodology and reliability. A partial solution to this problem was presented in and made publicly accessible through the ProCKSI webserver. A more complete list of currently available and freely distributed structural alignment software can be found in structural alignment software.
Properties of some structural alignment servers and software packages are summarized and tested with examples at Structural Alignment Tools in Proteopedia.Org.
See also
- Multiple sequence alignment
- List of sequence alignment software
- Sequence alignment
- Structural Classification of Proteins
References
- Zhang Y, Skolnick J. (2005). "The protein structure prediction problem could be solved using the current PDB library". Proc Natl Acad Sci USA. 102 (4): 1029–34. doi:10.1073/pnas.0407152101. PMC 545829. PMID 15653774.
- ^ Zemla A. (2003). "LGA — A Method for Finding 3-D Similarities in Protein Structures". Nucleic Acids Research. 31 (13): 3370–3374. doi:10.1093/nar/gkg571. PMC 168977. PMID 12824330.
- Godzik A (1996). "The structural alignment between two proteins: Is there a unique answer?". Protein science : a publication of the Protein Society. 5 (7): 1325–38. doi:10.1002/pro.5560050711. PMC 2143456. PMID 8819165.
- Martin ACR (1982). "Rapid Comparison of Protein Structures". Acta Cryst. A38 (6): 871–873. doi:10.1107/S0567739482001806.
- Theobald DL, Wuttke DS (2006). "Empirical Bayes hierarchical models for regularizing maximum likelihood estimation in the matrix Gaussian Procrustes problem". Proceedings of the National Academy of Sciences. 103 (49): 18521–18527. doi:10.1073/pnas.0508445103.
- Theobald DL, Wuttke DS (2006). "THESEUS: Maximum likelihood superpositioning and analysis of macromolecular structures". Bioinformatics. 22 (17): 2171–2172. doi:10.1093/bioinformatics/btl332. PMC 2584349. PMID 16777907.
- Diederichs K. (1995). "Structural superposition of proteins with unknown alignment and detection of topological similarity using a six-dimensional search algorithm". Proteins. 23 (2): 187–95. doi:10.1002/prot.340230208. PMID 8592700.
- Maiti R, Van Domselaar GH, Zhang H, Wishart DS (2004). "SuperPose: a simple server for sophisticated structural superposition". Nucleic Acids Res. 32 (Web Server issue): W590–4. doi:10.1093/nar/gkh477. PMC 441615. PMID 15215457.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Lathrop RH. (1994). "The protein threading problem with sequence amino acid interaction preferences is NP-complete". Protein Eng. 7 (9): 1059–68. doi:10.1093/protein/7.9.1059. PMID 7831276.
- Wang L, Jiang T. (1994) On the complexity of multiple sequence alignment). "On the complexity of multiple sequence alignment". J Comput Biol. 1 (4): 337–348. doi:10.1089/cmb.1994.1.337. PMID 8790475.
{{cite journal}}: Check date values in:|year=(help)CS1 maint: year (link) - Siew N, Elofsson A, Rychlewsk L and Fischer D (2000). "MaxSub: an automated measure for the assessment of protein structure prediction quality". Bioinformatics. 16 (9): 776–85. doi:10.1093/bioinformatics/16.9.776. PMID 11108700.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Poleksic A (2009). "Algorithms for optimal protein structure alignment". Bioinformatics. 25 (21): 2751–2756. doi:10.1093/bioinformatics/btp530. PMID 19734152.
- Kolodny R, Linial N. (2004). "Approximate protein structural alignment in polynomial time". PNAS. 101 (33): 12201–12206. doi:10.1073/pnas.0404383101. PMC 514457. PMID 15304646.
- Martinez L, Andreani, R, Martinez, JM. (2007). "Convergent algorithms for protein structural alignment". BMC Bioinformatics. 8: 306. doi:10.1186/1471-2105-8-306. PMC 1995224. PMID 17714583.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Mount DM. (2004). Bioinformatics: Sequence and Genome Analysis 2nd ed. Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY ISBN 0879697121
- Holm L, Sander C (1996). "Mapping the protein universe". Science. 273 (5275): 595–603. doi:10.1126/science.273.5275.595. PMID 8662544.
- ^ Shindyalov, I.N. (1998). "Protein structure alignment by incremental combinatorial extension (CE) of the optimal path". Protein Engineering. 11 (9): 739–747. doi:10.1093/protein/11.9.739. PMID 9796821.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - Prlic A, Bliven S, Rose PW, Bluhm WF, Bizon C, Godzik A, Bourne PE (2010). "Pre-calculated protein structure alignments at the RCSB PDB website". Bioinformatics (Oxford, England). 26 (23): 2983–2985. doi:10.1093/bioinformatics/btq572. PMC 3003546. PMID 20937596.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Guerler A, Knapp E.W. (2008). "Novel protein folds and their nonsequential structural analogs". Protein Science. 17 (8): 1374–82. doi:10.1110/ps.035469.108. PMC 2492825. PMID 18583523.
- Guerler, A; Moll, S; Weber, M; Meyer, H; Cordes, F (2008). "Selection and flexible optimization of binding modes from conformation ensembles". Bio Systems. 92 (1): 42–8. doi:10.1016/j.biosystems.2007.11.004. PMID 18241979.
- ^ Ortiz, A. R., C. E. Strauss, and O. Olmea (2002). "Mammoth (matching molecular models obtained from theory): An automated method for model comparison". Protein Sci. 11 (11): 2606–2621. doi:10.1110/ps.0215902. PMC 2373724. PMID 12381844.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Qiu, J.; Hue, M.; Ben-Hur, A.; Vert, J.-P.; Noble, W. S. (2007). "A structural alignment kernel for protein structures". Bioinformatics. 23 (9): 1090–8. doi:10.1093/bioinformatics/btl642. PMID 17234638.
- WCG Status Update. Predictions of structures of all unknown small domains in 150 genones. nyu.edu
- Kedem, K., Chew, L., and Elber, R. (1999). "Unit-vector RMS (URMS) as a tool to analyze molecular dynamics trajectories". Proteins. 37 (4): 554–64. doi:10.1002/(SICI)1097-0134(19991201)37:4<554::AID-PROT6>3.0.CO;2-1. PMID 10651271.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Lupyan, D., A. Leo-Macias, and A. R. Ortiz (2005, August). "A new progressive-iterative algorithm for multiple structure alignment". Bioinformatics. 21 (15): 3255–3263. doi:10.1093/bioinformatics/bti527. PMID 15941743.
{{cite journal}}: Check date values in:|year=(help)CS1 maint: multiple names: authors list (link) CS1 maint: year (link) - Konc J., Janežič D. (2010). "ProBiS algorithm for detection of structurally similar protein binding sites by local structural alignment". Bioinformatics. 26 (9): 1160–8. doi:10.1093/bioinformatics/btq100. PMC 2859123. PMID 20305268.
- Mosca, Schneider TR; Schneider, T. R. (2008). "RAPIDO: A web server for the alignment of protein structures in the presence of conformational changes". Nucleic Acids Research. 36 (Web Server issue): W42–6. doi:10.1093/nar/gkn197. PMC 2447786. PMID 18460546.
- Schneider, TR (2002). "A genetic algorithm for the identification of conformationally invariant regions in protein molecules". Acta crystallographica D. 58 (Pt 2): 195–208. PMID 11807243.
- Teichert, F. Bastolla, U. Porto, M. (2007). "SABERTOOTH: Protein structure comparison based on a vectorial structure representation". BMC Bioinformatics. 8: 425. doi:10.1186/1471-2105-8-425. PMC 2257979. PMID 17974011.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - Taylor WR, Flores TP, Orengo CA. (1994). "Multiple protein structure alignment". Protein Sci. 3 (10): 1858–70. doi:10.1002/pro.5560031025. PMC 2142613. PMID 7849601.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Orengo CA, Michie AD, Jones S, Jones DT, Swindells MB, Thornton JM. (1997). "CATH: A hierarchical classification of protein domain structures". Structure. 5 (8): 1093–1108. doi:10.1016/S0969-2126(97)00260-8. PMID 9309224.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Yang Y, Zhan J, Zhao H, Zhou Y. (2012). "A new size-independent score for pairwise protein structure alignment and its application to structure classification and nucleic-acid binding prediction". Proteins. 80 (8): n/a. doi:10.1002/prot.24100. PMC 3393833. PMID 22522696.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Ilyin lab. similarity "Protein structure alignment by TOPOFIT".
{{cite web}}: Check|url=value (help) - Ilyin, V; Abyzov, A; Leslin, CM (2004). "Structural alignment of proteins by a novel TOPOFIT method, as a superimposition of common volumes at a topomax point". Protein Science. 13 (7): 1865–1874. doi:10.1110/ps.04672604. PMC 2279929. PMID 15215530.
- Krissinel, E; Henrick, K (2004). "Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions". Acta crystallographica. Section D, Biological crystallography. 60 (Pt 12 Pt 1): 2256–68. doi:10.1107/S0907444904026460. PMID 15572779.
- ^ Zhang Y, Skolnick J. (2005) TM–align: A protein structure alignment algorithm based on TM–score). "TM-align: A protein structure alignment algorithm based on the TM-score". Nucleic Acids Research. 33 (7): 2302–2309. doi:10.1093/nar/gki524. PMC 1084323. PMID 15849316.
{{cite journal}}: Check date values in:|year=(help)CS1 maint: year (link) - Zhang Y, Skolnick J. (2004) Scoring function for automated assessment of protein structure template quality). "Scoring function for automated assessment of protein structure template quality". Proteins. 57 (4): 702–710. doi:10.1002/prot.20264. PMID 15476259.
{{cite journal}}: Check date values in:|year=(help)CS1 maint: year (link) - ^ Barthel D., Hirst J.D., Blazewicz J., Burke E.K. and Krasnogor N. (2007). "ProCKSI: a decision support system for Protein (Structure) Comparison, Knowledge, Similarity and Information". BMC Bioinformatics. 8: 416. doi:10.1186/1471-2105-8-416. PMC 2222653. PMID 17963510.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - Torarinsson E, Sawera M, Havgaard JH, Fredholm M, Gorodkin J. (2006). "Torarinsson E, Sawera M, Havgaard JH, Fredholm M, Gorodkin J". Genome Res. 16 (7): 885–9. doi:10.1101/gr.5226606. PMC 1484455. PMID 16751343.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Hoksza D, Svozil D. (2012). "Efficient RNA pairwise structure comparison by SETTER method". Bioinformatics. 28 (14): 1858–1864. doi:10.1093/bioinformatics/bts301. PMID 22611129.
- Cech P, Svozil D, Hoksza D. (2012). "SETTER: web server for RNA structure comparison". Nucleic Acids Research. 40 (W1): W42 – W48. doi:10.1093/nar/gks560.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Havgaard JH, Lyngso RB, Stormo GD, Gorodkin J. (2005). "Pairwise local structural alignment of RNA sequences with sequence similarity less than 40%". Bioinformatics. 21 (9): 1815–24. doi:10.1093/bioinformatics/bti279. PMID 15657094.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Mathews DH, Turner DH. (2006). "Prediction of RNA secondary structure by free energy minimization". Curr Opin Struct Biol. 16 (3): 270–8. doi:10.1016/j.sbi.2006.05.010. PMID 16713706.
Cite error: A list-defined reference named "wcg_yeast" is not used in the content (see the help page).
Further reading
- Bourne PE, Shindyalov IN. (2003): Structure Comparison and Alignment. In: Bourne, P.E., Weissig, H. (Eds): Structural Bioinformatics. Hoboken NJ: Wiley-Liss. ISBN 0-471-20200-2
- Yuan X, Bystroff C. (2004) "Non-sequential Structure-based Alignments Reveal Topology-independent Core Packing Arrangements in Proteins", Bioinformatics. Nov 5, 2004
- Jung J, Lee B (2000). "Protein structure alignment using environmental profiles". Protein Eng. 13: 535–543.
- Ye Y, Godzik A (2005). "Multiple flexible structure alignment using partial order graphs". Bioinformatics. 21 (10): 2362–2369.
- Sippl M, Wiederstein M (2008). "A note on difficult structure alignment problems". Bioinformatics. 24 (3): 426–427.
Category: